9 Calibration Measures
This page is a work in progress and minor changes will be made over time.
Calibration measures evaluate the ‘average’ quality of survival distribution predictions. This chapter is kept relatively short as the literature in this area is scarce (Rahman et al. 2017), this is likely due to the meaning of calibration being unclear in a survival context (Van Houwelingen 2000). However the meaning of calibration is better specified once specific metrics are introduced. As with other measure classes, only measures that can generalise beyond Cox PH models are included here but note that several calibration measures for re-calibrating PH models have been discussed in the literature (Demler, Paynter, and Cook 2015; Van Houwelingen 2000).
Calibration measures can be grouped (Andres et al. 2018) into those that evaluate distributions at a single time-point, ‘1-Calibration’ or ‘Point Calibration’ measures, and those that evaluate distributions at all time-points ‘distributional-calibration’ or ‘probabilistic calibration’ measures. A point-calibration measure will evaluate a function of the predicted distribution at a single time-point whereas a probabilistic measure evaluates the distribution over a range of time-points; in both cases the evaluated quantity is compared to the observed outcome, \((t, \delta)\).
9.1 Point Calibration
Point calibration measures can be further divided into metrics that evaluate calibration at a single time-point (by reduction) and measures that evaluate an entire distribution by only considering the event time. The difference may sound subtle but it affects conclusions that can be drawn. In the first case, a calibration measure can only draw conclusions at that one time-point, whereas the second case can draw conclusions about the calibration of the entire distribution. This is the same caveat as using prediction error curves for scoring rules (Section 10.3).
9.1.1 Calibration by Reduction
Point calibration measures are implicitly reduction methods as they use classification methods to evaluate a full distribution based on a single point only (Chapter 19). For example, given a predicted survival function \(\hat{S}\), one could calculate the survival function at a single time point, \(\hat{S}{\tau}\) and then use probabilistic classification calibration measures. Using this approach one may employ common calibration methods such as the Hosmer–Lemeshow test (Hosmer and Lemeshow 1980). Measuring calibration in this way can have significant drawbacks as a model may be well-calibrated at one time-point but poorly calibrated at all others (Haider et al. 2020). To mitigate this, one could perform the Hosmer–Lemeshow test (or other applicable tests) multiple times with multiple testing correction at many (or all possible) time points, however this would be less efficient and more difficult to interpret than other measures discussed in this chapter.
9.1.2 Houwelingen’s \(\alpha\)
As opposed to evaluating distributions at one or more arbitrary time points, one could instead evaluate distribution predictions at meaningful times. van Houwelingen proposed several measures (Van Houwelingen 2000) for calibration but only one generalises to all probabilistic survival models, termed here ‘Houwelingen’s \(\alpha\)’. The measure assesses if the model correctly estimates the theoretical ‘true’ cumulative hazard function of the underlying data generating process, \(H = \hat{H}\).
The statistic is derived by noting the closely related nature of survival analysis and counting processes, and exploiting the fact that the sum of the cumulative hazard function is an estimate for the number of events in a given time-period (Hosmer Jr, Lemeshow, and May 2011). As this result may seem surprising, below is a short experiment using \(\textsf{R}\) that demonstrates how the sum of the cumulative hazard estimated by a Kaplan-Meier estimator is identical to the number of randomly simulated deaths in a dataset:
set.seed(42)
library(survival)
event = rbinom(100, 1, 0.7)
times = runif(100)
H = survfit(Surv(times, event) ~ 1)$cumhaz
c("Deaths" = sum(event), "Sum H" = sum(H))
#> Deaths Sum H
#> 66 66Houwelingen’s \(\alpha\) is then defined by substituting \(H\) for the observed total number of deaths and summing over all predictions:
\[ H_\alpha(\boldsymbol{\delta}, \hat{\mathbf{H}}, \mathbf{t}) = \frac{\sum_i \delta_i}{\sum_i \hat{H}_i(t_i)} \]
with standard error \(SE(H_\alpha) = \exp(1/\sqrt{\sum_i \delta_i})\). A model is well-calibrated with respect to \(H_\alpha\) if \(H_\alpha = 1\).
The next metrics we look at evaluate models across a spectrum of points to assess calibration over time.
9.2 Probabilistic Calibration
Calibration over a range of time points may be assessed quantitatively or qualitatively, with graphical methods often favoured. Graphical methods compare the average predicted distribution to the expected distribution, which can be estimated with the Kaplan-Meier curve, discussed next.
9.2.1 Kaplan-Meier Comparison
The simplest graphical comparison compares the average predicted survival curve to the Kaplan-Meier curve estimated on the testing data. Let \(\hat{S}_1,...,\hat{S}_m\) be predicted survival functions, then the average predicted survival function is the mixture: \(\bar{\hat{S}} = \frac{1}{m} \sum^{m}_{i = 1} \hat{S}_i(\tau)\). This estimate can be plotted next to the Kaplan-Meier estimate of the survival distribution in a test dataset (i.e., the true data for model evaluation), allowing for visual comparison of how closely these curves align. An example is given in Figure 9.1, a Cox model (CPH), random survival forest, and relative risk tree, are all compared to the Kaplan-Meier estimator. This figure highlights the advantages and disadvantages of this method. The relative risk tree is clearly poorly calibrated as it increasingly diverges from the Kaplan-Meier. In contrast, the Cox model and random forest cannot be directly compared to one another, as both models frequently overlap with each other and the Kaplan-Meier estimator. Hence it is possible to say that the Cox and forests models are better calibrated than the risk tree, however it is not possible to say which of those two is better calibrated and whether their distance from the Kaplan-Meier is significant or not at a given time (when not clearly overlapping).
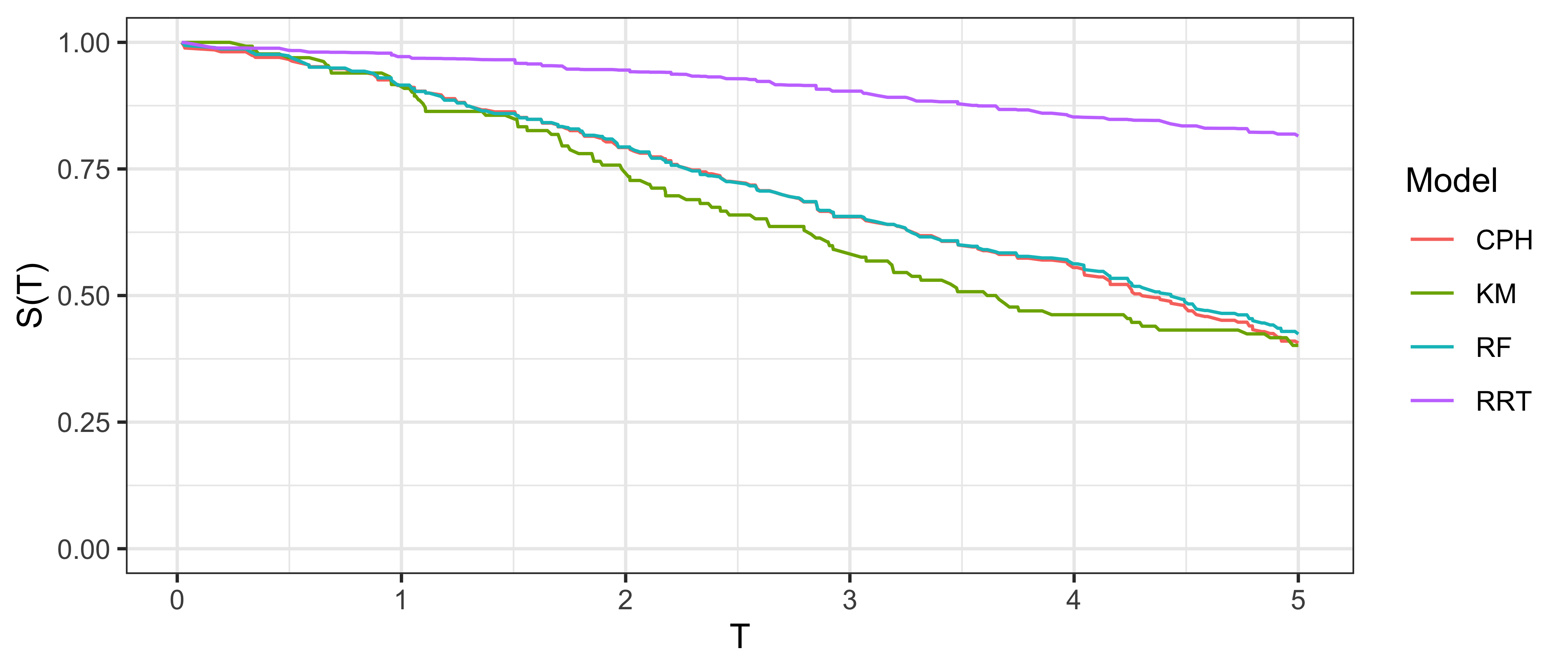
This method is useful for making broad statements such as “model X is clearly better calibrated than model Y” or “model X appears to make average predictions close to the Kaplan-Meier estimate”, but that is the limit in terms of useful conclusions. One could refine this method for more fine-grained information by instead using relative risk predictions to create ‘risk groups’ that can be plotted against a stratified Kaplan-Meier, however this method is harder to interpret and adds even more subjectivity around how many risk groups to create and how to create them (Royston and Altman 2013). The next measure we consider includes a graphical method as well as a quantitative interpretation.
9.2.2 D-Calibration
D-Calibration (Andres et al. 2018; Haider et al. 2020) evaluates a model’s calibration by assessing if the predicted survival distributions follow the Uniform distribution as expected, which is motivated by the result that for any random variable \(X\) it follows \(S_X(x) \sim \mathcal{U}(0,1)\). This can be tested using a \(\chi^2\) test-statistic:
\[ \chi^2 := \sum_{i=1}^n \frac{(O_i - E_i)^2}{E_i} \]
where \(O_1,...,O_n\) is the observed number of events in \(n\) groups and \(E_1,...,E_n\) is the expected number of events.
To utilise this test, the \([0,1]\) codomain of \(S_i\) is cut into \(B\) disjoint contiguous intervals (‘bins’) over the full range \([0,1]\). Let \(m\) be the total number of observations, then assuming a discrete uniform distribution as the theoretical distribution, the expected number of events in each bin is \(E_i = m/B\) (as the probability of an observation falling into each bin is equal).
The observations in the \(i\)th bin, \(b_i\), are defined by the set:
\[ b_i := \{j = 1,\ldots,m : \lceil \hat{S}_i(t_j)B \rceil = i\} \]
where \(j = 1,\ldots,m\) are the indices of the observations, \(\hat{S}_i\) are observed (i.e., predicted) survival functions, \(t_i\) are observed (i.e., the ground truth) outcome times, and \(\lceil \cdot \rceil\) is the ceiling function. The observed number of events in \(b_i\) is then the number of observations in that set: \(O_i = |b_i|\).
The D-Calibration measure, or \(\chi^2\) statistic, is now defined by,
\[ D_{\chi^2}(\hat{\mathbf{S}}, \mathbf{t}) := \frac{\sum^B_{i = 1} (O_i - \frac{m}{B})^2}{m/B} \]
where \(\hat{\mathbf{S}} = (\hat{S}_1 \ \hat{S}_2 \cdots \hat{S}_m)^\top\) and \(\mathbf{t}= (t_1 \ t_2 \cdots t_m)^\top\).
This measure has several useful properties. Firstly, one can test the null hypothesis that a model is ‘D-calibrated’ by deriving a \(p\)-value from comparison to \(\chi^2_{B-1}\). Secondly, \(D_{\chi^2}\) tends to zero as a model is increasingly well-calibrated, hence the measure can be used for model comparison. Finally, the theory lends itself to an intuitive graphical calibration method as a D-calibrated model implies:
\[ p = \frac{\sum_i \mathbb{I}(T_i \leq \hat{F}_i^{-1}(p))}{m} \]
where \(p\) is some value in \([0,1]\), \(\hat{F}_i^{-1}\) is the \(i\)th predicted inverse cumulative distribution function, and \(m\) is again the number of observations. In words, the number of events occurring at or before each quantile should be equal to the quantile itself, for example 50% of events should occur before their predicted median survival time. Therefore, one can plot \(p\) on the x-axis and the right hand side of the above equation on the y-axis. A D-calibrated model should result in a straight line on \(x = y\). This is visualised in Figure 9.2 for the same models as in Figure 9.1. This figure supports the previous findings that the relative risk tree is poorly calibrated in contrast to the Cox model and random forest but again no direct comparison between the latter models is possible.
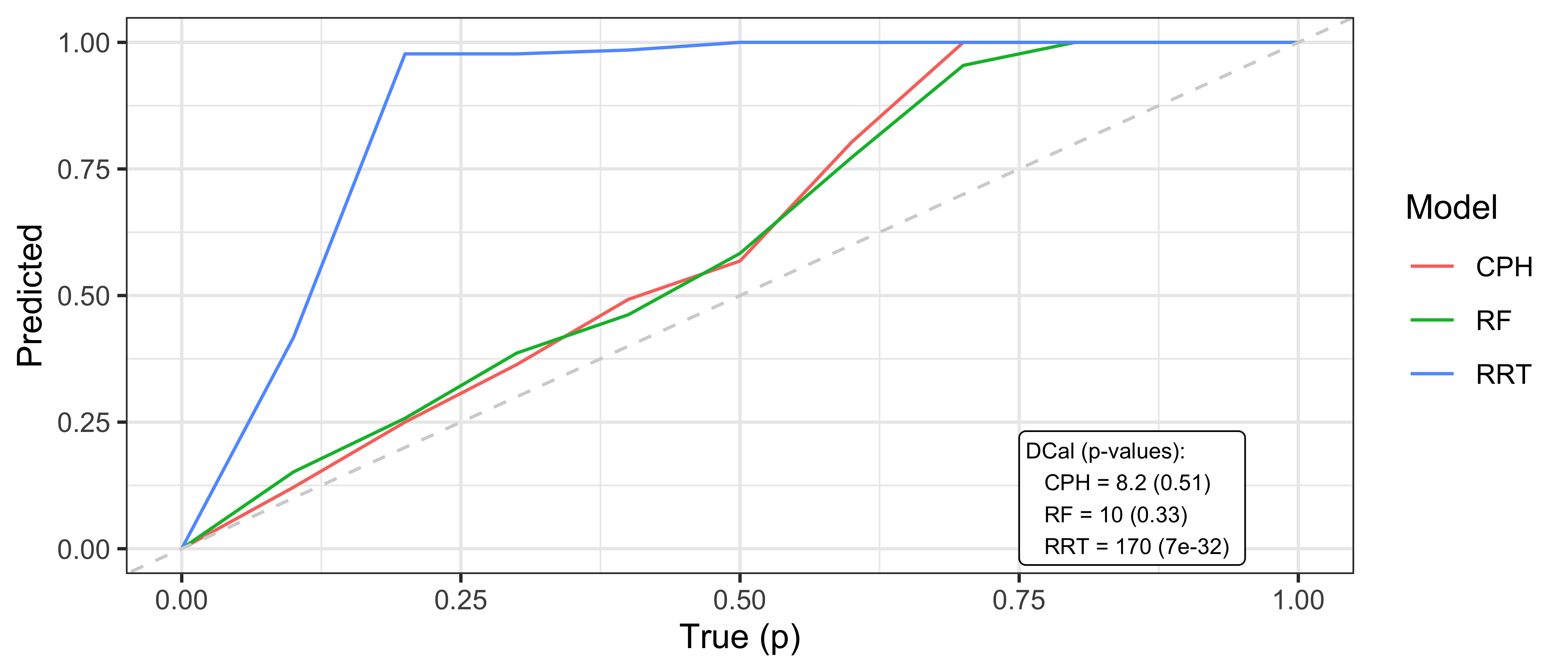
Whilst D-calibration has the same problems as the Kaplan-Meier method with respect to visual comparison, at least in this case there are quantities to help draw more concrete solutions. For the models in Figure 9.2, it is clear that the relative risk tree is not D-calibrated with \(p<0.01\) indicating the null hypothesis of D-calibration, i.e., the predicted quantiles not following a Discrete Uniform distribution, can be comfortably rejected. Whilst the D-calibration for the Cox model is smaller than that of the random forest, the difference is unlikely to be significant, as is seen in the overlapping curves in the figure.
The next chapter will look at scoring rules, which provides a more concrete method to analytically compare the predicted distributions from survival models.