2 Machine Learning
This page is a work in progress and minor changes will be made over time.
This chapter covers core concepts in machine learning. This is not intended as a comprehensive introduction and does not cover mathematical theory nor how to run machine learning models using software. Instead, the focus is on introducing important concepts and to provide basic intuition for a general machine learning workflow. This includes the concept of a machine learning task, data splitting (resampling), model training and prediction, evaluation, and model comparison. Recommendations for more comprehensive introductions are given at the end of this chapter, including books that cover practical implementation in different programming languages.
2.1 Basic workflow
This book focuses on supervised learning, in which predictions are made for outcomes based on data with observed dependent and independent variables. For example, predicting someone’s height is a supervised learning problem as data can be collected for features (independent variables) such as age and sex, and an observable outcome (dependent variable), which is height. Alternatives to supervised learning include unsupervised learning, semi-supervised learning, and reinforcement learning. This book is primarily concerned with predictive survival analysis, i.e., making future predictions based on (partially) observed survival outcomes, which falls naturally within the supervised learning domain.
The basic machine learning workflow is represented in Figure 2.1. Data is split into training and test datasets. A learner is selected and is trained on the training data, inducing a fitted model. The features from the test data are passed to the model which makes predictions for the unseen outcomes (Box 1). The outcomes from the test data are passed to a chosen measure with the predictions, which evaluates the performance of the model (Box 2). The process of repeating this procedure to test different training and test data is called resampling and running multiple resampling experiments with different models is called benchmarking. All these concepts will be explained in this chapter.

2.2 Tasks
A machine learning task is the specification of the mathematical problem that is to be solved by a given algorithm. For example, “predict the height of a male, 13 year old child”, is a machine learning task. Tasks are derived from datasets and one dataset can give rise to many tasks across any machine learning domain. The dataset described by columns: ‘age’, ‘weight’, ‘height’, ‘sex’, ‘diagnosis’, ‘time of death’, ‘clinician notes’, could give rise to any of the following tasks (and more):
- Predict age from weight, height, and sex - supervised regression task
- Predict sex from age and diagnosis - supervised classification task
- Predict time of death from all other features - supervised survival task
- Categorise observations into clusters - unsupervised clustering
- Learn to speak like a clinician depending on client diagnosis - natural language processing, likely with reinforcement learning
As this book is focused on supervised learning, only the first three of these is covered in this chapter and beyond. The specification of a task is vital for interpreting predictions from a model and its subsequent performance. This is particularly true when separating between determinisitc and probabilistic predictions, as discussed later in the chapter.
Formally, let \(\mathbf{x}\in \mathcal{X}\subseteq \mathbb{R}^{n \times p}\) be a matrix with \(p\) features for \(n\) observations and let \(y \in \mathcal{Y}\) be a vector of labels (or outcomes or targets) for all observations. A dataset is then given by \(\mathcal{D}= ((\mathbf{x}_1, y_1) , . . . , (\mathbf{x}_n, y_n))\) where it is assumed \(\mathcal{D}\stackrel{i.i.d.}\sim(\mathbb{P}_{xy})^n\) for some unknown distribution \(\mathbb{P}\).
A machine learning task is the problem of learning the unknown function \(f : \mathcal{X}\rightarrow \mathcal{Y}\) where \(\mathcal{Y}\) specifies the nature of the task, for example classification, regression, or survival.
2.2.1 Regression
Regression tasks make continuous predictions, for example someone’s height. Regression may be deterministic, in which case a single continuous value is predicted, or probabilistic, where a probability distribution over the Reals is predicted. For example, predicting an individual’s height as 165cm would be a deterministic regression prediction, whereas predicting their height follows a \(\mathcal{N}(165, 2)\) distribution would be probabilistic.
Formally, a deterministic regression task is specified by \(f_{Rd} : \mathcal{X}\rightarrow \mathcal{Y}\subseteq \mathbb{R}^n\), and a probabilistic regression task by \(f_{Rp} : \mathcal{X}\rightarrow \mathcal{S}\) where \(\mathcal{S}\subset \operatorname{Distr}(\mathcal{Y})\) and \(\operatorname{Distr}(\mathcal{Y})\) is the space of distributions over \(\mathcal{Y}\).
In machine learning, deterministic regression is much more common than probabilistic and hence the shorthand ‘regression’ is used to refer to deterministic regression (in contrast to statistical modeling, where regression usually implies probabilistic regression).
2.2.2 Classification
Classification tasks make discrete predictions, for example whether it will rain, snow, or be sunny tomorrow. Similarly to regression, predictions may be deterministic or probabilistic. Deterministic classification predicts which category an observation falls into, whereas probabilistic classification predicts the probability of an observation falling into each category. Predicting it will rain tomorrow is a deterministic prediction whereas predicting \(\hat{p}(rain) = 0.6; \hat{p}(snow) = 0.1; \hat{p}(sunny) = 0.3\) is probabilistic.
Formally, a deterministic classification task is given by \(f_{Cd} : \mathcal{X}\rightarrow \mathcal{Y}\subseteq \mathbb{N}_0\), and a probabilistic classification task as \(f_{Cp} : \mathcal{X}\rightarrow \mathcal{Y}\subseteq [0,1]^k\) where \(k\) is the number of categories an observation may fall into. Practically this latter prediction is estimation of the probability mass function \(\hat{p}_Y(y) = P(Y = y)\). If only two categories are possible, these reduce to the binary classification tasks: \(f_{Bd}: \mathcal{X}\rightarrow \{0, 1\}\) and \(f_{Bp}: \mathcal{X}\rightarrow [0, 1]\) for deterministic and probabilistic binary classification respectively.
Note that in the probabilistic binary case it is common to write the task as predicting \([0,1]\) not \([0,1]^2\) as the classes are mutually exclusive. The class for which probabilities are predicted is referred to as the positive class, and the other as the negative class.
2.3 Training and predicting
The terms algorithm, learner, and model are often conflated in machine learning. A learner is a description of a learning algorithm, prediction algorithm, parameters, and hyperparameters. The learning algorithm is a mathematical strategy to estimate the unknown mapping from features to outcome as represented by a task, \(f: \mathcal{X}\rightarrow \mathcal{Y}\). During training, data, \(\mathcal{D}\), is fed into the learning algorithm and induces the model \(\hat{f}\). Whereas the learner defines the framework for training and prediction, the model is the specific instantiation of this framework after training on data.
After training the model, new data, \(\mathbf{x}^*\), can be fed to the prediction algorithm, which is a mathematical strategy that uses the model to make predictions \(\hat{\mathbf{y}}= \hat{f}(\mathbf{x}^*)\). Algorithms can vary from simple linear equations with coefficients to estimate, to complex iterative procedures that differ considerably between training and predicting.
Algorithms usually involve parameters and hyperparameters. Parameters are learned from data whereas hyperparameters are set beforehand to guide the algorithms. Model parameters (or weights), \(\boldsymbol{\theta}\), are coefficients to be estimated during model training. Hyperparameters, \(\boldsymbol{\lambda}\), control how the algorithms are run but are not directly updated by them. Hyperparameters can be mathematical, for example the learning rate in a gradient boosting machine (Chapter 13), or structural, for example the depth of a decision tree (Chapter 11). The number of hyperparameters usually increases with learner complexity and affects its predictive performance. Often hyperparameters need to be tuned (Section 2.5) instead of manually set. Computationally, storing \((\hat{\boldsymbol{\theta}}, \boldsymbol{\lambda})\) is sufficient to recreate any trained model.
2.4 Evaluating and benchmarking
To understand if a model is ‘good’, its predictions are evaluated with a loss function. Loss functions assign a score to the discrepancy between predictions and true values, \(L: \mathcal{Y}\times \mathcal{Y}\rightarrow \bar{\mathbb{R}}\). Given (unseen) real-world data, \((\mathbf{X}^*, \mathbf{y}^*)\), and a trained model, \(\hat{f}\), the loss is given by \(L(\hat{f}(\mathbf{X}^*), \mathbf{y}^*) = L(\hat{\mathbf{y}}, \mathbf{y}^*)\). For a model to be useful, it should perform well in general, meaning its generalization error should be low. The generalization error refers to the model’s performance on new data, rather than just the data encountered during training and development.
A model should only be used to make predictions if its generalization error was estimated to be acceptable for a given context. If a model were to be trained and evaluated on the same data, the resulting loss, known as the training error, would be an overoptimistic estimate of the true generalization error (James et al. 2013). This occurs as the model is making predictions for data it has already ‘seen’ and the loss is therefore not evaluating the model’s ability to generalize to new, unseen data. Another reason for data splitting is to help mitigate model overfitting, which occurs when models learn noise in the training data leading them to perform worse on new data. This can be avoided using data splitting methods, feature engineering (manually or using built-in machine learning methods) to remove noisy features, and early stopping which stops training a model once a certain performance has been reached.
Estimation of the generalization error requires data splitting, which is the process of splitting available data, \(\mathcal{D}\), into training data, \(\mathcal{D}_{train}\subset \mathcal{D}\), and testing data, \(\mathcal{D}_{test}= \mathcal{D}\setminus \mathcal{D}_{train}\).
The simplest method to estimate the generalization error is to use holdout resampling, which is the process of partitioning the data into one training dataset and one testing dataset, with the model trained on the former and predictions made for the latter. Using 2/3 of the data for training and 1/3 for testing is a common splitting ratio (Kohavi 1995). For independent and identically distributed (iid) data, it is generally best practice to partition the data randomly. This ensures that any potential patterns or information encoded in the ordering of the data are removed, as such patterns are unlikely to generalize to new, unseen data. For example, in clinical datasets, the order in which patients enter a study might inadvertently encode latent information such as which doctor was on duty at the time, which could theoretically influence patient outcomes. As this information is not explicitly captured in measured features, it is unlikely to hold predictive value for future patients. Random splitting breaks any spurious associations between the order of data and the outcomes.
When data is not iid, for example spatially correlated or time-series data, then random splitting may not be advisable, see Hornung et al. (2023) for an overview of evaluation strategies in non-standard settings.
Holdout resampling is a quick method to estimate the generalization error, and is particular useful when very large datasets are available. However, hold-out resampling has a very high variance for small datasets and there is no guarantee that evaluating the model on one hold-out split is indicative of real-world performance.
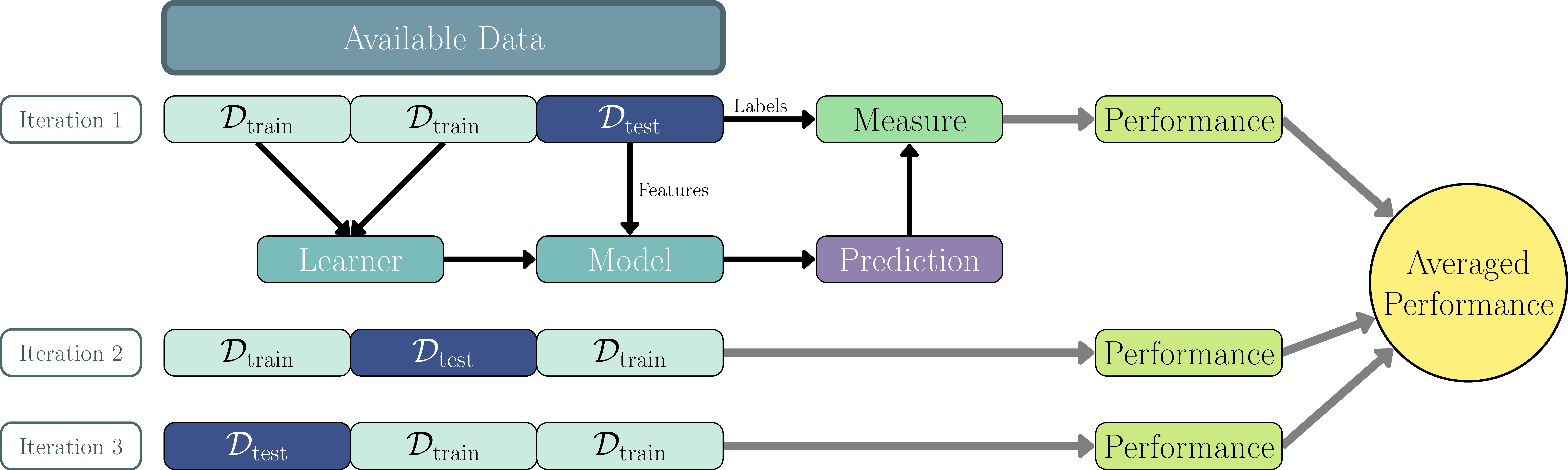
\(k\)-fold cross-validation (CV) can be used as a more robust method to better estimate the generalization error (Hastie, Tibshirani, and Friedman 2001). \(k\)-fold CV partitions the data into \(k\) subsets, called folds. The training data comprises of \(k-1\) of the folds and the remaining one is used for testing and evaluation. This is repeated \(k\) times until each of the folds has been used exactly once as the testing data. The performance from each fold is averaged into a final performance estimate (Figure 2.2). It is common to use \(k = 5\) or \(k = 10\) (Breiman and Spector 1992; Kohavi 1995). This process can be repeated multiple times (repeated \(k\)-fold CV) and/or \(k\) can even be set to \(n\), which is known as leave-one-out cross-validation.
Cross-validation can also be stratified, which ensures that a variable of interest will have the same distribution in each fold as in the original data. This is important, and often recommended, in survival analysis to ensure that the proportion of censoring in each fold is representative of the full dataset (Casalicchio and Burk 2024; Herrmann et al. 2021).
Repeating resampling experiments with multiple models is referred to as a benchmark experiment. A benchmark experiment compares models by evaluating their performance on identical data, which means the same resampling strategy and folds should be used for all models. Determining if one model is actually better than another is a surprisingly complex topic (Benavoli et al. 2017; Demšar 2006; Dietterich 1998; Nadeau and Bengio 2003) and is out of scope for this book, instead any benchmark experiments performed in this book are purely for illustrative reasons and no results are expected to generalize outside of these experiments. A common heuristic is to suggest one model outperforms another if it performs better across all folds in a repeated cross-validation benchmark experiment, however this is just a heuristic and without robust hypothesis testing results should be interpreted with caution.
2.5 Hyperparameter Optimization
Section 2.3 introduced model hyperparameters, which control how training and prediction algorithms are run. Setting hyperparameters is a critical part of model fitting and can significantly change model performance. Tuning is the process of using internal benchmark experiments to automatically select the optimal hyper-parameter configuration. For example, the depth of trees, \(m_r\) in a random forest (Chapter 11) is a potential hyperparameter to tune. This hyperparameter may be tuned over a range of values, say \([1, 15]\) or over a discrete subset, say \(\{1, 5, 15\}\), for now assume the latter. Three random forests with \(1\), \(5\), and \(15\) tree depth respectively are compared in a benchmark experiment. The depth that results in the model with the optimal performance is then selected for the hyperparameter value going forward. Nested resampling is a common method to reduce bias that could occur from using overlapping data for tuning, training, or testing (Simon 2007). Nested resampling is the process of resampling the training set again for tuning and then the optimal model is refit on the entire training data (Figure 2.3).
